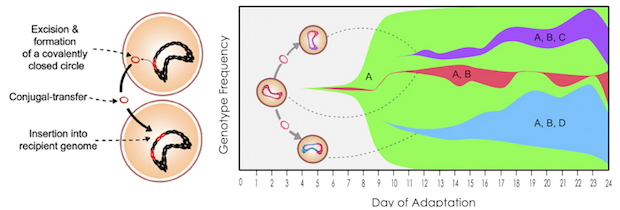
This chart shows how quickly resistance to tigecycline, an antibiotic, spread through a population of Enterococcus faecalis bacteria that was exposed to increasing tigecycline concentrations over the course of 24 days in experiments at Rice University. A mutation in the ribosomal S10 protein quickly reached 100 percent frequency in the population (A). Next, a mutation leading to TetM overexpression occurred in Tn916 (B) and subsequently spread through the population by increased Tn916 horizontal gene transfers (C and D). The dashed lines show instances when Tn916 conjugation likely occurred. Click image to see a larger version. Graphic by Kathryn Beabout
Rice University lab identifies mutations that allow bacteria to resist antibiotics
Rice University scientists are developing strategies to keep germs from evolving resistance to antibiotics by heading them off at the pass.
The Rice lab of biochemist Yousif Shamoo identified a genetic mechanism that allows bacteria to develop resistance while simultaneously and quickly spreading the capability to others in a population.
“This is really a double whammy,” Shamoo said. “Our finding that these bacteria become more antibiotic-resistant while at the same time spreading their resistance more efficiently was really surprising and worrying.”
The researchers hope this knowledge will help predict when and how bacterial strains are likely to develop resistance to future antibiotics and perhaps act to halt — or at least slow — the process. The research appeared in the journal Molecular Biology and Evolution.

Kathryn Beabout, lead author of a Rice University study on antibiotic resistance. The study identified a genetic mechanism that allows bacteria to develop resistance while simultaneously and quickly spreading the capability to others in a population. The researchers' goal is to develop strategies to keep germs from evolving such resistance. Photo by Jeff Fitlow
Antibiotic resistance is responsible for hundreds of thousands of infections acquired in American hospitals, according to the Centers for Disease Control and Prevention. These infections kill thousands of patients. While progress is being made to control microbes that spread infection, the overriding concern remains that drugs developed to kill germs will ultimately stop working.
Until now, the only effective way to keep antibiotics from losing their potency has been to use them sparingly, said Kathryn Beabout, a Rice graduate student and lead author of the new paper.
“The best you can do is try to manage when you use the antibiotic,” she said. “But our idea is that if we can predict how resistance is going to emerge, we can come up with strategies to use antibiotics in a more intelligent way.”
The lab used experimental evolution to study a specific combination of bacteria and an antibiotic that had not been in common contact. The bacteria of interest was Enterococcus faecalis, found in the gastrointestinal tract. The antibiotic was tigecycline, a highly effective but sparingly used derivative of tetracycline. The goal was to see how horizontal gene transfer – the means by which cells pass along favorable mutations – would work in the presence of the antibiotic.
It worked quite well, they found.
That was due mostly to a chunk of mutant DNA known as Tn916, a transposon that can change its position along the genome, duplicate itself and pass to other cells in a process known as parasexuality, the exchange of genetic material between cells. Tn916 carries the tetracycline-resistance gene called tetM, which has been found in many pathogens, according to the researchers.
Without tigecycline, Tn916 moves only infrequently, as about one in 120,000 bacteria transfer their resistance to another bacteria. But in the presence of the antibiotic, Tn916 movement rose to one in 50 bacteria, due to a mutation that also causes overproduction of tetM. The resistance mechanism required the presence of two mutations, to Tn916 and to a gene that encodes the ribosomal S10 protein, both of which could be easily identified through pre- and post-experiment gene sequencing.
‘We are seeing both resistance and an increase in the frequency at which resistance is able to transfer from cell to cell and move around genomes. That’s definitely worrisome.’ — Kathryn Beabout
“Tetracyclines bind to the cell’s ribosome and prevent it from making proteins,” Beabout said. “TetM is a protein that comes in and kicks tetracyclines off and frees the ribosome, but it doesn’t usually work against tigecycline. We didn’t expect to see it emerge.”
In their experiments, the researchers discovered the mutations led to the production of large amounts of tetM proteins. “When you have an abundance of tetM, they’re able to have an effect against tigecycline,” Beabout said. “What’s really interesting is that tetM is on a conjugative transposon (Tn916), which is a DNA element that’s able to move around the genome and can be transferred to other cells.
“An additional effect of this tetM overexpression is that the Tn916 transposon moves more,” she said. “It passes from cell to cell more. So we are seeing both resistance and an increase in the frequency at which resistance is able to transfer from cell to cell and move around genomes. That’s definitely worrisome.”
The lab allowed colonies of E. faecalis to grow rapidly in bioreactors for 19 and 24 days. The results showed the bacteria were remarkably proficient at picking up the resistance gene. “All the cells at the beginning had one copy of the transposon, and throughout the experiment they started acquiring additional copies. The copy number of the transposon was increasing very rapidly.”
Shamoo hopes the research leads to drugs that inhibit resistance mechanisms and preserve the effectiveness of new antibiotics.
“Our lab does a form of evolutionary reconnaissance into how bacteria will become resistant in the future,” Shamoo said. “The pharmaceutical industry and other labs can use this information to develop drugs to stay ahead of the pathogens.”
Co-authors of the paper are postdoctoral associate Troy Hammerstrom, undergraduate Tim Wang and faculty fellow Gerda Saxer, all at Rice; and postdoctoral fellow Minny Bhatty and Peter Christie, a professor of microbiology and molecular genetics at the University of Texas Medical School at Houston. Shamoo is a professor of biochemistry and cell biology and Rice’s vice provost for research.
The U.S. Army Edgewood Chemical Biological Center, the National Institute of Allergy and Infectious Diseases and the Defense Threat Reduction Agency supported the research.


Leave a Reply