Texas Medical Center-based team unravels how loops form in genome
A research team based in Houston’s Texas Medical Center has found that the proteins that turn genes on by forming loops in human chromosomes work like the sliding plastic adjusters on a grade-schooler’s backpack. This discovery could provide new clues about genetic diseases and allow researchers to reprogram cells by directly modifying the loops in genomes.
The study, which appears online this week in the Proceedings of the National Academy of Sciences, is by the same team that published the first high-resolution 3-D maps showing how the human genome folds inside the nucleus of a cell. The multi-institutional group includes researchers from Baylor College of Medicine, Rice University, Stanford University and the Broad Institute.
http://youtu.be/Sv2EGB73U4k
Every human cell contains a genome, a linear string of DNA. Sequences of DNA bases spell out genes, much like letters spell out words. For decades, scientists have known that genes that lie far apart on the string can activate one another by looping back and coming into contact during genome folding. Last year, the team showed that it was possible to map the positions of these loops, and the researchers created the first atlas of loops in the human genome. But the group couldn’t explain how the loops were forming.
“For months, we had no idea what our data really meant,” said senior author Erez Lieberman Aiden, a geneticist and computer scientist with joint appointments at Baylor and Rice. “Then one day, we realized that we’d been carrying the solution around — literally, on our back — for decades!”
The human genome contains more than 20,000 genes. In any given cell, only a fraction of these are active, and this fraction determines the cell’s function: whether it will become a hard-pumping heart cell, a body-defending immune cell or a metastatic cancer cell. Many genes are activated by loops, and it is impossible to understand gene activation without knowing how loops form, Aiden said.
Aiden, who is also a senior investigator at Rice’s Center for Theoretical Biological Physics, said the researchers found that a set of proteins acts like the plastic slider, sometimes called a tri-glide, that adjusts a backpack strap.
“The mechanism that makes this possible can be explained to any kindergartener with a backpack,” said study co-first author Adrian Sanborn, a graduate student in the Aiden lab and at Stanford University. “The protein complex that forms DNA loops appears to operate like the plastic slider that is used to adjust the length of the straps: it lands on DNA and takes up slack to form a loop.”
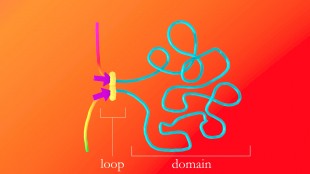
The protein complex that forms DNA loops takes up slack by feeding DNA through from either direction until it hits the DNA keyword, or motif, which acts like a brake. The motifs must point in the correct direction; if they point away from the protein complex, it will not recognize them and will keep taking up slack. Credit: A. Sanborn, S. Rao
Aiden, assistant professor of genetics at Baylor and of computer science and computational and applied mathematics at Rice, said Sanborn and study co-first author Suhas Rao showed that they could combine the tri-glide model with mathematics and high-performance computation to predict how a genome will fold. The team confirmed their predictions by making tiny modifications in a cell’s genome and showing that the mutations changed the folding pattern exactly as expected. Rao likened the result to a new form of genome surgery: a procedure that can modify how a genome is folded by design and with extraordinary precision.
“We found that changing even one letter in the genetic code was enough to modify the folding of millions of other letters,” said Rao, a graduate student in the Aiden lab and at Stanford University. “What was stunning was that once we understood how the loops were forming, the results of these changes became extremely predictable.”
Sanborn said the discovery also explains a puzzling pattern that the team noticed when it published its original atlas of loops.
“DNA encodes information, and you can think of each DNA base pair as a letter and of certain sequences of letters as words,” he said. “In our data, we noticed that when particular keywords appeared, a loop would form. But the loop would only form if the two keywords were pointing at one another. For example, if one side of the loop read K-E-Y-W-O-R-D, the other would be D-R-O-W-Y-E-K.”
“That’s an incredibly strange thing because these words can be millions of letters apart, and the genome is flexible at that scale,” said Sanborn. “If I were a protein, and I wanted to bring these two words together, it’s difficult to envision why the way that the keyword is pointing would matter. This simple fact was a crucial clue.”
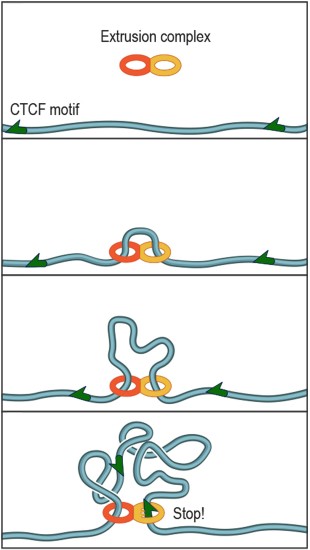
The protein complex that forms DNA loops operates like the plastic slider that is used to adjust the length of backpack straps. The complex lands on DNA and takes up slack to form a loop. Researchers discovered that a specific DNA keyword called a "motif" -- a string of fewer than 20 genetic letters that causes the DNA to bind a protein called CTCF -- lies at each end of nearly all loops. Credit: A. Sanborn, S. Rao
That clue eventually led the team to the tri-glide theory, but not before a series of false starts. First, the team tested models based on fractal packing, but they proved mathematically that such packing could not explain the data. Next, the researchers tested a model of DNA folding where tension along the DNA chain caused it to condense like an elastic band, but this model also did not fit the data.
Eventually, they hit on the tri-glide model. The basic idea is that the tri-glide protein complex lands on the genome and pulls the strand from each side so that a loop forms in the middle — just like the loop someone might make if they wanted to tighten a backpack strap.
“The strand just keeps feeding through and feeding through from each direction until it hits the keyword, which acts like a brake,” said Rao, a student in the Aiden lab and at Stanford University. “So it’s not so much that the keywords need to point at one another; it’s that they need to point at the tri-glide complex because the complex won’t recognize them if they point the other way. To the complex, they would look upside-down.”
Aiden said that one of the most astonishing implications of the new model is that loops on different chromosomes tend not to become entangled.
“In the old model, scientists thought that a loop formed when two bits of the genome wiggled around and then met inside the cell nucleus,” Aiden said. “But this process would lead to interweaving loops and highly entangled chromosomes. This is a big problem if you need those chromosomes to separate again when the cell divides.
“The tri-glide takes care of that,” he said. “Even in a big pile of backpacks, you can use your tri-glide to make a loop without any risk of entanglement.”
Aiden said the discoveries were possible, in part, because of Rice’s new PowerOmics supercomputer, which allowed his team to analyze more 3-D folding data than was previously possible. The high-performance computer, an IBM POWER8 system that is customized with a cluster of NVIDIA graphical processing units, allowed Aiden’s group to run analyses in a few hours that would previously have taken several days or even weeks, he said.
Additional study co-authors include Su-Chen Huang, Neva C. Durand, Miriam H. Huntley, Andrew Jewett, Ivan D. Bochkov, Dharmaraj Chinnappan, Ashok Cutkosky, Jian Li, Kristopher P. Geeting, Doug McKenna and Elena K. Stamenova of the Center for Genome Architecture at Baylor; and Eric Lander, Andreas Gnirke, and Alexandre Melnikov of the Broad Institute, a collaboration between Harvard University and the Massachusetts Institute of Technology. Aiden, Sanborn and Li are also with the Center for Theoretical Biological Physics at Rice University. McKenna is also with Mathemaesthetics Inc. in Boulder, Colo.
The research was supported by the Welch Foundation, IBM, Nvidia, the National Science Foundation, the National Institutes of Health, the Cancer Prevention and Research Institute of Texas and the McNair Medical Institute. Rice’s PowerOmics high-performance computer cluster is managed by the Office of Information Technology’s Center for Research Computing in a partnership with the Ken Kennedy Institute for Information Technology.



This is an exciting announcement. Congrats to Erez, Adrian, Suhos and the broader team that made this happen.