NEWS RELEASE
Editor’s note: Links to high-resolution images for download appear at the end of this release.
Jeff Falk
713-348-6775
jfalk@rice.edu
Mike Williams
713-348-6728
mikewilliams@rice.edu
Cancer cells’ plasticity makes them harder to stop
Rice U. team links genomic, metabolic processes in cells’ evasive strategies as they metastasize
HOUSTON – (Feb. 7, 2019) – When metastatic cancer cells need to avoid a threat, they simply reprogram themselves. Rice University scientists are beginning to get a handle on how they survive hostile environments.
Members of Rice’s Center for Theoretical Biological Physics (CTBP) and cancer metabolism researchers at Baylor College of Medicine have created a basic framework of how cancer cells — whether in tumors or as single cells — adapt when their attempts to metastasize are blocked by drugs or the body’s immune system. Understanding the cells’ strategies could someday help scientists design therapies that keep them in check.
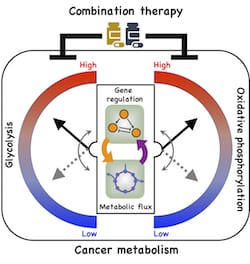
Their model shows a direct connection between gene regulation and metabolic pathways and how cancer cells take advantage of it to adapt to hostile environments, a process known as metabolic plasticity.
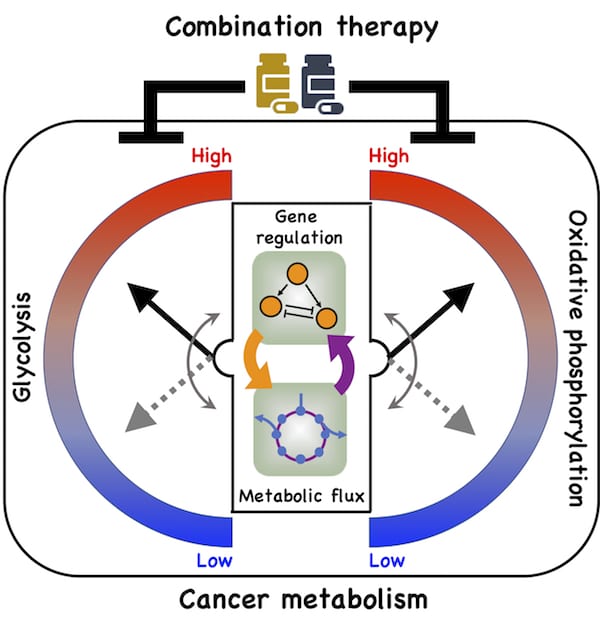
A new model by Rice University researchers details a direct connection between gene expression and metabolism and how cancer cells take advantage of it to adapt to hostile environments, a process known as metabolic plasticity. Illustration by Dongya Jia
In particular, the team led by physicists Herbert Levine and José Onuchic and postdoctoral fellow Dongya Jia looked at oxidative phosphorylation (OXPHOS) and glycolysis, metabolic processes that provide cells with the energy and chemical building blocks they need to proliferate.
From their model, they detailed for the first time a direct association between the activities of two protein players, AMP-activated protein kinase (AMPK) and hypoxia-inducible factor-1 (HIF-1), the master regulators of OXPHOS and glycolysis, respectively, with the activities of three major metabolic pathways: glucose oxidation, glycolysis and fatty acid oxidation.
Their theoretical model was experimentally supported by Baylor cancer mitochondrial metabolism researchers led by Dr. Benny Abraham Kaipparettu.
The new study appears in the Proceedings of the National Academy of Sciences.
“A lot of early cancer papers focus on the Warburg effect, when cancer cells primarily use glycolysis even in the presence of oxygen,” Onuchic said. “This is true, but it’s not like cancer cells give up on other mechanisms. The more aggressive they become, the more they are able to use any available choice to acquire energy. Our model shows how that’s possible.”
“Only recently have people paid attention to OXPHOS,” Jia added. “But they don’t really understand how cancer cells regulate these two metabolic phenotypes. We want to know how cancer cells orchestrate them. Since there is an extensive cross-talk between gene regulation and metabolic pathways, we think it’s necessary to simultaneously look at these two different aspects of cancer metabolism.”

Rice University researchers — from left, Dongya Jia, Herbert Levine and José Onuchic — detail a direct connection between gene expression and metabolism and how cancer cells take advantage of it to adapt to hostile environments.
The researchers said their model helped the team hone in on critical processes that traditional genome-scale metabolic models might miss. “We start with simple models where we can figure out completely what’s going on, and then we add details to that scaffold without losing the basic understanding of how the system’s working,” Levine said.
Jia’s mathematical model details connections that allow cancer cells to adopt three stable metabolic states. One is a glycolytic state, characterized by high activity of HIF-1 and high activity of the glycolytic pathway. The second is an OXPHOS state, characterized by high activity of AMPK and high activity of such OXPHOS pathways as glucose oxidation and fatty acid oxidation.
The third is a hybrid metabolic state characterized by high activity of AMPK and HIF-1 and of the glycolysis and OXPHOS pathways. The Rice model revealed the presence of both HIF-1 and AMPK can lead to the hybrid state that is difficult for current cancer therapies to address.
The researchers also found the hybrid metabolic state can be promoted by the stabilization of HIF-1 and the elevated production rate of mitochondrial reactive oxygen species (ROS) in cancer cells relative to normal cells. ROS are chemically active molecules that are important to signaling but at high levels can damage cells.
Kaipparettu’s Baylor team backed up the theory using gene expression data from breast cancer patients and metastatic triple negative breast cancer experimental models. Experimental evidence showed that repressing glycolytic activity in the cells activated AMPK and enhanced OXPHOS. The reverse was also true. But a combination of inhibitors that attacked both glycolysis and OXPHOS successfully eliminated the cells’ metabolic plasticity.
“We’re trying to push the field of metabolic modeling towards more flexibility, allowing for the decision-making processes we see in cells,” Levine said. “And here we’re coupling genes to metabolism in a way that’s rather novel.
“It’s still a limited view of all the metabolic pathways,” he said. “There are yet other possibilities that are not included in our model. We eventually need to tell a more complete story to really know what’s happening.”
Co-authors of the paper are former Rice postdoctoral researcher Mingyang Lu, an assistant professor at The Jackson Laboratory, Bar Harbor, Maine; postdoctoral associates Kwang Hwa Jung and Jun Hyoung Park of Baylor; and Rice alumnus Linglin Yu. Levine is an adjunct professor of bioengineering at Rice and a University Distinguished Professor at Northeastern University. Onuchic is the Harry C. and Olga K. Wiess Chair of Physics, a professor of physics and astronomy, of chemistry and of biochemistry and cell biology and co-director of the CTBP.
The research was supported by the National Science Foundation; National Cancer Institute; The Jackson Laboratory; the National Institute of General Medical Sciences; the Department of Defense; the Collaborative Faculty Research Investment Program, the Dan L. Duncan Comprehensive Cancer Center and the Charles A. Sammons Cancer Center, all at Baylor; and the Breast Cancer Research Foundation.
-30-
Read the abstract at www.pnas.org/cgi/doi/10.1073/pnas.1816391116.
Follow Rice News and Media Relations via Twitter @RiceUNews.
Related materials:
José Onuchic:https://chemistry.rice.edu/people/jose-onuchic
Herbert Levine: http://www.bioe.neu.edu/people/levine-herbert
Benny Abraham Kaipparettu: https://www.bcm.edu/people/view/benny-kaipparettu-ph-d/b1915357-ffed-11e2-be68-080027880ca6
Center for Theoretical Biological Physics: https://ctbp.rice.edu/
Wiess School of Natural Sciences: https://naturalsciences.rice.edu
Images for download:
https://news2.rice.edu/files/2019/02/0201_CANCER-1-WEB-uwdkgi.jpg
A new model by Rice University researchers details a direct connection between gene expression and metabolism and how cancer cells take advantage of it to adapt to hostile environments, a process known as metabolic plasticity. (Credit: Illlustration by Dongya Jia/Rice University)
https://news2.rice.edu/files/2019/02/0201_CANCER-2-WEB-11ezu6r.jpg
Rice University researchers — from left, Dongya Jia, Herbert Levine and José Onuchic — detail a direct connection between gene expression and metabolism and how cancer cells take advantage of it to adapt to hostile environments. (Credit: Rice University)
Located on a 300-acre forested campus in Houston, Rice University is consistently ranked among the nation’s top 20 universities by U.S. News & World Report. Rice has highly respected schools of Architecture, Business, Continuing Studies, Engineering, Humanities, Music, Natural Sciences and Social Sciences and is home to the Baker Institute for Public Policy. With 3,962 undergraduates and 3,027 graduate students, Rice’s undergraduate student-to-faculty ratio is just under 6-to-1. Its residential college system builds close-knit communities and lifelong friendships, just one reason why Rice is ranked No. 1 for lots of race/class interaction and No. 2 for quality of life by the Princeton Review. Rice is also rated as a best value among private universities by Kiplinger’s Personal Finance. To read “What they’re saying about Rice,” go to http://tinyurl.com/RiceUniversityoverview.