Rice, Rutgers scientists test early proteins for potential as building blocks in synthetic biology
Electrons are common currency in biological systems, supplying the power to live. Scientists at Rice and Rutgers universities have found a way to harness them through a very old metabolic transaction.
Their study in the Proceedings of the National Academy of Sciences details the successful manipulation of simple ferredoxin proteins that shuttle electrons from here to there in cells, and in the process shows they may have been doing so since the start of life on Earth.
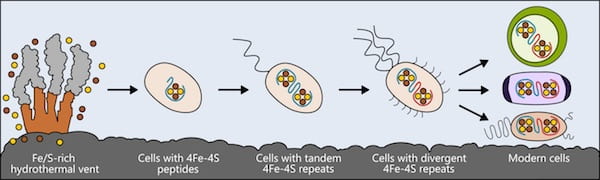
Life may have arisen near hydrothermal vents rich in iron and sulfur. The earliest cells incorporated these elements into small peptides, which became the first and simplest ferredoxins — proteins that shuttle electrons within the cell — to support metabolism. As cells evolved, ferredoxins mutated into more complex forms. The ferredoxins in modern bacteria, plant and animal cells are all derived from that simple ancestor. Illustration by Ian Campbell
Ferredoxins are small proteins that contain clusters of iron and sulfur. They are biological capacitors that get charged as they accept electrons from one molecule and discharged as they deliver them to other molecules. A method to control how these molecular “alligator clips” function could have broad implications for synthetic biology, including processes that manipulate microbes to produce industrial chemicals.
The new study led by Rutgers biophysicist Paul Falkowski and computational biologist Vikas Nanda resides at the crossroads of evolutionary biology, biochemistry, geology and synthetic biology. The latter is thanks to the participation of Rice bioscientists Joff Silberg and George Bennett and graduate student Ian Campbell, who joined the project to see if the Rutgers-designed proteins might be robust enough to thrive in living bacteria.

Scientists at Rice and Rutgers universities have manipulated simple ferredoxin proteins that shuttle electrons in cells. The discovery could impact synthetic biology processes used to program microbes that produce biofuels and other industrial products. Photo by Jeff Fitlow
Rice already had a connection to Falkowski and Nanda through the Rutgers-based ENIGMA Project, a NASA-funded exploration of the origin of proteins on Earth and, by extension, the possibility of such evolution on other planets. Through ENIGMA, Falkowski and Nanda revealed in a 2018 paper their design of a small, synthetic ferredoxin they believe could have been an evolutionary building block for life 4 billion years ago.
Concurrent with the Rutgers find, Silberg’s lab created synthetic proteins able to switch the flow of electrons on and off in biological systems and proved their abilities in Escherichia coli bacteria. Testing Rutgers’ proteins in bacteria seemed like a natural collaboration.
“When we started to talk with Rutgers, the proteins they’ve been building in test tubes really aligned with the cellular assays that we developed for monitoring electron flow,” Silberg said. “We told them we could study whether these proteins not only hold an electron in a test tube but are also robust enough to support a biological process that requires many cycles of electron transfer.”
To test the theory, they programmed E. coli to produce several designed variations of a ferredoxin dimer. These proteins had a symmetrical inner core that contained iron-sulfur clusters and modular outer terminals that served as binding sites and as donors or acceptors in an electron transfer circuit. “It’s an ancient alligator clip, effectively,” Silberg said. “Its job is to grab an electron from one thing and give it to another. That’s all it does. But the cool thing is the symmetry of it.”

Rice University graduate student Ian Campbell pulls a vial of ferredoxin proteins from cold storage. The iron and sulfur proteins, believed to be present at the start of life on Earth, facilitate the transfer of energy in cells. The Rice experiments showed synthetic biologists may use them to control electron transfer in cells. Photo by Jeff Fitlow
Depending on the dimers’ configurations and the level of donor and acceptor targets, they found the proteins were easily able to transfer electrons and enhance cell growth.
Campbell suggested the protein’s modular nature lends itself to the creation of libraries of synthetic electron shuttles that can be customized to target specific molecules in a cell and to control the rate of electron transfer.
“I think the next step is to push it back even further in evolutionary time and make recreations of the ancient monomer, half of what’s been made so far,” he said. “That hasn’t been done yet. Once we start to understand how each individual half functions, maybe we can swap out one half for another that binds a different metal, perhaps, or another kind of electron carrier.”
“Once you start to understand the rules of design, you can build synthetic pathways that control exactly where the electrons flow,” Silberg said. “It becomes much more of a science, not an empirical, ‘Let’s try something that kind of works.'”

Rice University synthetic biologist Joff Silberg, left, and graduate student Ian Campbell. Photo by Jeff Fitlow
One direct beneficiary of the discovery may be Bennett’s Rice lab, which genetically engineers microbes to produce biofuels and industrial chemicals.
The lab has previously shown ferredoxins can be used to rewire electron flow within Clostridium acetobutylicum, an anaerobic organism used in biofuel production. By introducing a non-native ferredoxin oxidoreductase, a protein that charges ferredoxins, the microbe was reprogrammed to produce a higher proportion of commercially valuable butanol. “The development of ferredoxins optimized for specific partners would better enable direction of metabolism toward desired products like butanol,” Bennett said.
While NASA’s concerns are the size of a planet and Silberg’s interests are at a somewhat smaller scale, he considers them equally important.
“They’re thinking about the evolution of biomolecules and the evolution of minerals,” he said. “A lot of minerals were driven by biological processes, so they want to know how the first proteins relate to the first minerals: If you find one, what does that mean for the other?
“But while they’re going back in time to what they believe are the most ancient proteins, we want to use those proteins to design more things,” he said.
Authors of the paper include postdoctoral researchers Andrew Mutter and Saroj Poudel and staff scientist Alexei Tyryshkin of Rutgers. Falkowski is a distinguished professor of Earth and planetary sciences and marine and coastal sciences and Nanda an associate professor of biochemistry and molecular biology at Rutgers. Bennett is the E. Dell Butcher Professor of Biochemistry and Cell Biology and Silberg the Stewart Memorial Professor of Biochemistry at Rice.
Supporting the research were the Gordon and Betty Moore Foundation, the Department of Energy, NASA, the Bennett L. Smith Endowment and the Rutgers University Institute of Earth, Ocean and Atmospheric Science Fellowship Program.

