SUMMARY: Rice University synthetic biologists design transcriptional circuits that allow single-cell microbes to form networks that spur collective action, even in large communities. The work could lead to engineered microbes that treat conditions in gut microbiomes or communicate with bioelectronics.
NEWS RELEASE
Jeff Falk
713-348-6775
jfalk@rice.edu
Mike Williams
713-348-6728
mikewilliams@rice.edu
Synthetic cells make long-distance calls
Rice scientists’ circuits help bacteria quickly pass signals to an entire community
The search for effective biological tools is a marathon, not a sprint, even when the distances are on the microscale. A discovery at Rice University on how engineered communities of cells communicate is a long step in the right direction.
The Rice lab of synthetic biologist Matthew Bennett has designed a set of transcriptional circuits that, when added to (and expressed by) the genomes of single-cell microbes, allows them to quickly form a network of local interactions to spur collective action, even in large communities.

In the video at left, Matthew Bennett describes two colonies of bacteria engineered to coordinate their activities. Video courtesy of the Bennett Lab and Nature Chemical Biology
Research published in Nature Chemical Biology shows engineered strains of Escherichia coli transmitting signals down a bacteria-filled corridor and coordinating their actions. The ability to do so could lead to engineered microbes that treat conditions in gut microbiomes or communicate with bioelectronics.
“Cells often use chemical signals to communicate and relay information to each other,” Bennett said. “However, chemical signals have a limited range. After they leave the cell, they diffuse through whatever medium the cells are in, and that can only go so far.
“In this study we looked at a previous system we built that uses two different strains and different types of communication between them to study how, once we increase the size of the colony containing these strains, it would react,” he said.
The evidence appears not only in a video from the lab that shows groups of microbes pulsing as they signal each other across the span of the experiment, but also in mathematical models by frequent collaborators Jae Kyoung Kim, a professor of mathematics at the Korea Advanced Institute of Science and Technology, and Krešimir Josić, a professor of mathematics at the University of Houston and an adjunct professor of biosciences at Rice.
The microbes were modified to express proteins that activated positive and negative feedback loops. To characterize the effect of the modifications, the researchers split them into four sets of either all positive, all negative, or combined positive and negative feedback loops.
They were then able to see that positive loops that activated signals were the most effective in facilitating communications. “We found you need the positive feedback loop in order to synchronize gene expression in spatially extended systems in which cells can’t directly communicate with one another.”
That’s not to say the negative circuits, or repressors, did nothing. “You need negative feedback in order to create and stabilize the oscillations,” Bennett said. “It makes the system more robust to perturbations in the environment.”
Bennett said the cells had no problem communicating across small microfluidic chambers in previous experiments. “There, the diffusion of the signaling molecule is very fast, and basically global,” he said. “All the cells can talk to each other because it’s such a confined space.
“In our new system, that’s just not true,” Bennett said. “Cells can only talk to their close-by neighbors and have no way to communicate with those at the other end of the colony. Despite this, we found the oscillations generated by the circuits we put into these two strains are still able to synchronize across space and time.”
In fact, signals they thought would take several hours to travel from one end of the chamber to the other caused synchronous oscillations almost immediately.
“We figured out some factors that are important for that, types of feedback loops that increase synchronization across these large extended colonies,” he said. “This is important because, as synthetic biologists move toward engineering larger, multicellular systems, we must be able to control not just what goes on within a single cell but also coordinate what happens in space and time within the whole population.”
The researchers found they had to give their cells some breathing room to communicate effectively, so they opened “doors” along the corridor. “We think that has to do with the stability of the strains,” Bennett said. “When you have two different types, they passively compete for space, jostling as they grow and divide.
“You can get local fluctuations where the cells may get pushed to one side. This creates instability and seriously complicates communication among the cells,” he said. “We found opening these traps stabilized the spatial makeup of the colony.”
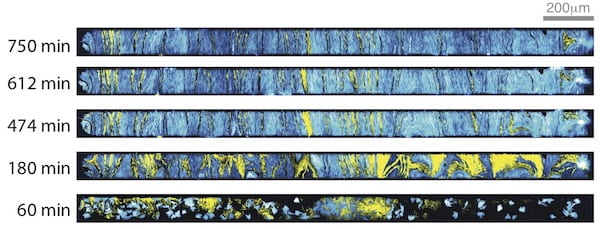
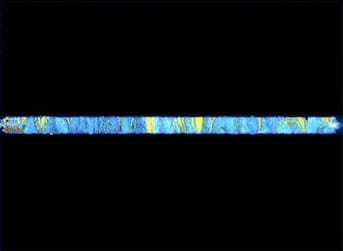
Representative fluorescence images of cells growing in the open microfluidic device developed at Rice University show how transcriptional circuits allow single-cell microbes to form networks that spur collective action, even in large communities. The spatial arrangement of two strains of color-coded cells eventually stabilizes. Courtesy of The Bennett Lab/Nature Chemical Biology
There’s much to do before such circuits can be deployed in the clinic, but Bennett sees a path forward. “Eventually we want to be able to build multicellular systems that do practical things,” he said. “Certainly, if we followed exactly what nature has done, we’d get a long way, because nature’s been very good at creating multicellular systems that do interesting things.
“But that doesn’t mean evolution has struck upon the only way to do things.”
Kim and Rice alumnus Ye Chen are co-lead authors of the paper. Co-authors are Josić, Rice postdoctoral researcher Andrew Hirning and Rice graduate student Razan Alnahhas. Bennett is an associate professor of biosciences and bioengineering.
The National Institutes of Health, the National Science Foundation, the Robert A. Welch Foundation, the Hamill Foundation, the National Research Foundation of Korea and the T.J. Park Science Fellowship of POSCO supported the research.
-30-
Read the abstract at https://www.nature.com/articles/s41589-019-0372-9
Follow Rice News and Media Relations via Twitter @RiceUNews.
Video:
(Credit: Courtesy of The Bennett Lab/Nature Chemical Biology)
Related materials:
The Bennett Lab: http://biodesign.rice.edu
Krešimir Josić: https://www.math.uh.edu/~josic/
Jae Kyoung Kim: http://mathsci.kaist.ac.kr/~jaekkim/
Rice Department of BioSciences: https://biosciences.rice.edu
Wiess School of Natural Sciences: https://naturalsciences.rice.edu
Image for download:
https://news2.rice.edu/files/2019/10/1014_GENE-1-WEB.jpg
Representative fluorescence images of cells growing in the open microfluidic device developed at Rice University show how transcriptional circuits allow single-cell microbes to form networks that spur collective action, even in large communities. The spatial arrangement of two strains of color-coded cells eventually stabilizes. (Credit: Courtesy of The Bennett Lab/Nature Chemical Biology)
Located on a 300-acre forested campus in Houston, Rice University is consistently ranked among the nation’s top 20 universities by U.S. News & World Report. Rice has highly respected schools of Architecture, Business, Continuing Studies, Engineering, Humanities, Music, Natural Sciences and Social Sciences and is home to the Baker Institute for Public Policy. With 3,962 undergraduates and 3,027 graduate students, Rice’s undergraduate student-to-faculty ratio is just under 6-to-1. Its residential college system builds close-knit communities and lifelong friendships, just one reason why Rice is ranked No. 1 for lots of race/class interaction and No. 4 for quality of life by the Princeton Review. Rice is also rated as a best value among private universities by Kiplinger’s Personal Finance.