Light-controlled genes could reveal how gut bacteria impact health
Baylor College of Medicine researcher Meng Wang had already shown that bacteria that make a metabolite called colanic acid (CA) could extend the lifespan of worms in her lab by as much as 50%, but her collaboration with Rice University synthetic biologist Jeffrey Tabor is providing tools to answer the bigger question of how the metabolite imparts longer life.
In a study published in eLife, Wang, Tabor and colleagues showed they could use different colors of light to turn gut bacteria genes on and off while the bacteria were in the intestines of worms. The work was made possible by an optogenetic control system Tabor has been developing for more than a decade.

Rice University’s Jeffrey Tabor is an associate professor of bioengineering and of biosciences and a faculty member of Rice’s Systems, Synthetic and Physical Biology Ph.D. Program. (Photo by Jeff Fitlow/Rice University)
“Meng’s group discovered that the CA compound could extend lifespan but they couldn’t say for sure whether this was a dietary ingredient that was being digested in the stomach or a metabolite that was being produced by bacteria in the intestines,” said Tabor, an associate professor of bioengineering and of biosciences at Rice. “We were able to restrict production of CA to the gut and show that it had a beneficial effect on cells in the intestines.”
For the experiments, Tabor’s lab engineered strains of E. coli to make CA when exposed to green, but not red, light. To make sure the bacteria worked properly, the team added genes to make different colors of fluorescent proteins that would show up brightly under a microscope. One color was always present, to make it easy to see where the bacteria were inside the worms, and a second color was made only when the bacteria were producing CA.
In collaboration with the Wang lab, Tabor’s lab kept the bacteria under a red light and fed them to worms, a species called Caenorhabditis elegans (C. elegans) that’s commonly used in life sciences. Researchers tracked the bacteria’s progress through the digestive tract and switched on the green light when they made it to the intestines.
“When exposed to green light, worms carrying this E. coli strain also lived longer. The stronger the light, the longer the lifespan,” said Wang, the Robert C. Fyfe Endowed Chair on Aging, a professor of molecular and human genetics at the Huffington Center on Aging at Baylor and a Howard Hughes Medical Institute investigator.
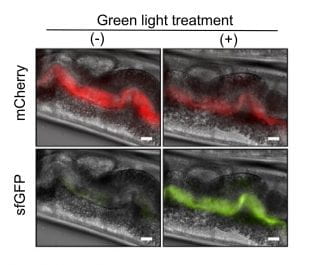
Light-responsive bacteria fed to worms are visible in images of the worms’ gastrointestinal tracts. Engineers programmed the bacteria to produce a red fluorescent protein called mCherry so they would be easy to see under a microscope. When exposed to green light, the bacteria also produce a green fluorescent protein called sfGFP, which causes them to glow green. When exposed to red light, they do not produce the green protein. Worms in the left column were treated with red light. Worms in the right column were treated with green light. (Image courtesy of Jeff Tabor/Rice University)
In the cells of C. elegans and other higher order life, from humans to yeast, specialized organelles called mitochondria supply most of the energy. Thousands of mitochondria work around the clock in each cell and maintain a dynamic balance between fission and fusion, but they become less efficient over time. As people and other organisms age, the dysfunction of mitochondria leads to functional decline in their cells.
In prior experiments with C. elegans, Wang and colleagues showed that CA can regulate the balance between mitochondrial fission and fusion in both intestinal and muscle cells to promote longevity. The worms typically live about three weeks, but Wang’s lab has shown that CA can extend their lives to 4.5 weeks — 50% longer than usual.
Tabor said this raises a host of questions. For instance, if CA is produced in the gut, do intestinal cells benefit first? Is the beneficial effect of CA related to its level? And most important, do the mitochondrial benefits spread throughout the body from the intestines?
In the eLife study, the researchers found that CA production in the gut directly improved mitochondrial function in intestinal cells in a short time. They did not find evidence of such direct, short-term mitochondrial benefits in the worms’ muscle cells. Thus, the longevity-promoting effect of CA starts from the gut and then spreads into other tissues over time.
“With our technology, we can use light to turn on CA production and watch the effect travel through the worm,” Tabor said.

Meng Wang is Baylor College of Medicine’s Robert C. Fyfe Endowed Chair on Aging, a professor of molecular and human genetics at Baylor’s Huffington Center on Aging and a Howard Hughes Medical Institute investigator. (Photo courtesy of Baylor College of Medicine)
He said the precision of the optogenetic technology could allow researchers to ask fundamental questions about gut metabolism.
“If you can control the timing and location of metabolite production with precision, you can think about experimental designs that show cause and effect,” he said.
Showing that gut bacteria directly impact health or disease would be a major achievement.
“We know gut bacteria affect many processes in our bodies,” Tabor said. “They’ve been linked to obesity, diabetes, anxiety, cancers, autoimmune diseases, heart disease and kidney disease. There’s been an explosion of studies measuring what bacteria you have when you have this illness or that illness, and it’s showing all kinds of correlations.”
But there is a big difference between showing correlation and causality, Tabor said.
“The goal, the thing you really want, is gut bacteria you can eat that will improve health or treat disease,” he said.
But it’s difficult for researchers to prove that molecules produced by gut bacteria cause disease or health. That’s partly because the gut is difficult to access experimentally, and it’s especially difficult to design experiments that show what is happening in specific locations inside the gut.
“The gut is a hard place to access, especially in large mammals,” Tabor said. “Our intestines are 28 feet long, and they’re very heterogeneous. The pH changes throughout and the bacteria change quite dramatically along the way. So do the tissues and what they’re doing, like the molecules they secrete.
“To answer questions about how gut bacteria influence our health, you need to be able to turn on genes in specific places and at particular times, like when an animal is young or when an animal wakes up in the morning,” he said. “You need that level of control to study pathways on their own turf, where they happen and how they happen.”
Because it uses light to trigger genes, optogenetics offers that level of control, Tabor said.
“To this point, light is really the only signal that has enough precision to turn on bacterial genes in the small versus the large intestine, for example, or during the day but not at night,” he said.
Tabor said he and Wang have discussed many ways they might use optogenetics to study aging.
“She’s found two dozen bacterial genes that can extend lifespan in C. elegans, and we don’t know how most of them work,” Tabor said. “The colanic acid genes are really intriguing, but there are many more that we’d like to turn on with light in the worm to figure out how they work. We can use the exact technique that we published in this paper to explore those new genes as well. And other people who are studying the microbiome can use it too. It’s a powerful tool for investigating how bacteria are benefiting our health.”
Tabor is a faculty member of Rice’s Systems, Synthetic and Physical Biology Ph.D. Program. Wang is also a member of Baylor’s Dan L Duncan Comprehensive Cancer Center.
Study co-authors include Lucas Hartsough, Matthew Kotlajich, John Tyler Lazar, Elena Musteata and Lauren Gambill of Rice, and Mooncheol Park, Bing Han and Chih-Chun Lin of Baylor. The research was supported by the National Institutes of Health, the National Aeronautics and Space Administration, the John S. Dunn Foundation and the Welch Foundation.

