Rice lab helps find useful protein that stops formaldehyde damage in methanol-eating microbes
Poison! That’s how some bacteria must react to the formaldehyde they themselves produce. They don’t want the toxin and have learned to stop it from doing too much damage before it’s too late.
Scientists now know how. Their discovery could help enhance other bacteria that break down plant matter to make biofuels, a process that also produces formaldehyde.
The Rice University laboratory of biochemist Yousif Shamoo, in collaboration with bioscientist and project leader Chris Marx at the University of Idaho, helped identify a protein that senses and binds to formaldehyde to tell a cell that toxic formaldehyde is building up inside them.
The research appears in PLOS Biology.
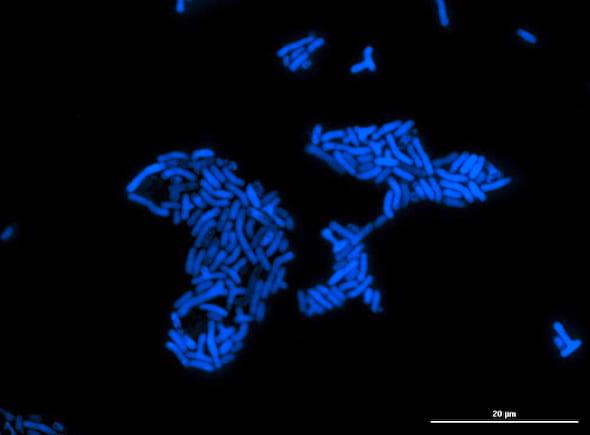
A microcolony of Methylorubrum extorquens that survives by consuming methanol also produces formaldehyde as a necessary, but toxic, byproduct. Scientists at the University of Idaho and Rice University discovered the microbe also produces a sensor protein, EfgA, that keeps the toxin in check to protect the organism. Photo by Nkrumah Grant/University of Idaho
Life on Earth is amazingly diverse. One group of bacteria called methylotrophs can use methanol or methane as a food source. The downside of using methanol is that the cells produce formaldehyde, which is highly toxic.
Marx and his Idaho colleagues, along with Shamoo, discovered EfgA (for “enhanced formaldehyde growth”) is a critical link in the metabolic chain that allows microorganisms to sense the amount of formaldehyde in their systems and act to protect themselves, basically acting as a “kill switch.” EfgA is the first known bacterial protein that senses the presence of a metabolic intermediate and halts protein translation.
Methanol, rather than glucose, is methylotrophs’ fuel of choice, but oxidizing it quickly produces free formaldehyde, Shamoo said. “Formaldehyde is extremely toxic,” he said. “How can a cell grow and yet keep from killing itself?”
The team discovered EfgA by evolving the methylotroph Methylorubrum extorquens to grow in the presence of formaldehyde. “This work has been a surprising outgrowth of a simple question: What does it take for cells to grow directly on formaldehyde?” Marx said. “Remarkably, they needed to break a sensor system rather than crank up detoxification.”

Yousif Shamoo
Shamoo and his lab then determined the structure and function of EfgA to provide the biochemical mechanism for how the protein can sense formaldehyde.
The authors believe EfgA interacts directly with ribosomes, the protein-producing factories in cells, to stop them from continuing to create new proteins that will be deformed upon reaction with formaldehyde.
The researchers then found when genes that encode EfgA were introduced into Escherichia coli bacteria, they too quickly developed protection against formaldehyde.
“It is remarkable that this one protein can confer protection not just in the methylotrophs but in other bacteria that do not use methanol as a food source,” Shamoo said. “This could be very useful in biotechnology applications.” Formaldehyde toxicity is a key hurdle in engineering E. coli that can use methanol as a sole carbon source, he said, and adding the EfgA gene could fix that.
Jannell Bazurto, an assistant professor of plant and microbial biology at the University of Minnesota and former postdoctoral researcher at Idaho, is lead author of the paper. Co-authors are former Rice research scientist Milya Davlieva and Dipti Nayak, Tomislav Ticak, Jessica Lee, Leah Lambert, Olivia Benski, Caleb Quates, Jill Johnson, Jagdish Suresh Patel and Marty Ytreberg of Idaho.
The National Institute of General Medical Sciences, the National Science Foundation, the BEACON Center for the Study of Evolution in Action, the Department of Organismic and Evolutionary Biology at Harvard University and a National Institutes of Health INBRE Undergraduate Research Fellowship supported the research.

