Rice, Northeastern computer models reveal new details of spike mechanism
The spike protein responsible for COVID-19 infection is like a loaded spring, with a catch.
New simulations may help researchers catch a break as they look for ways to defeat the SARS-CoV-2 virus.
Scientists at Rice University and Northeastern University, all part of the Rice-based Center for Theoretical Biological Physics (CTBP), have simulated the process by which the spike reconfigures itself to attach to and infect cells.
The catch comes in the form of glycans, sugar molecules that are attached to the “head” domain of the spike and enclose it while the fusion peptide “tails” go in search of a cell to invade. The cage formed by these steric (nonbinding but influential) glycans hold the head domain in check, giving the fusion peptides time to look around.
Once a host is found, the head and viral membrane are pulled toward the tail, fusing with the unfortunate cell.
“The glycans give the fusion peptides enough time to diffuse,” said Rice physicist José Onuchic, co-principal investigator and co-director of CTBP. “Finding a way to speed up the spike’s conformation may be one approach to keeping the peptides from fishing for a host cell.”
The research appears in the open-access journal eLife.
“The glycans serve the virus because they’re not recognized as an invader,” said Rice graduate student and lead author Esteban Dodero-Rojas. “They’re a shield that makes an immune response to the virus difficult.”
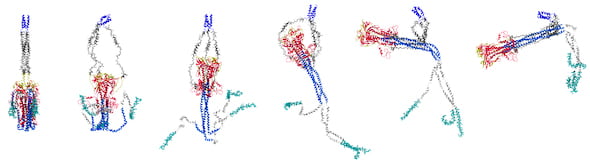
Simulations by scientists at the Rice University-based Center for Theoretical Biological Physics suggest how the SARS-CoV-2 spike infects cells. The illustration shows how the spike reconfigures itself in microseconds as it goes from pre- to post-fusion with target cells. The researchers suggest their work to reveal the mechanism by which the virus spreads could lead to new strategies to defeat COVID-19. (Credit: Esteban Dodero-Rojas/Rice University)
Co-principal investigator Paul Whitford, an associate professor at Northeastern and senior scientist at CTBP, said that while little can be done about the presence of glycans, revealing their significance in the spike’s mechanism could offer previously unknown opportunities to protect against COVID-19.
“Glycans are all over the place, and in principle, a glycan-free system would make the virus pretty ineffective — but introducing something that removes them in the cell would likely have major side effects,” he said. “However, the simulations show us the full conformational change of the spike, which basically has to turn inside out. This huge rearrangement reveals many more points where we could potentially target the system.
“Some of the elements are completely buried at the start and finish, but transiently, they’re accessible,” Whitford said.
A previous study involving the team, a collaboration with experimentalists at Rutgers University, used computational models to find hooks on the spike for an inhalable vaccine. The new study takes a more comprehensive look at how the spike changes to deliver its viral load to the cell.
In the model, the process starts when the spike 1 (S1) domain that covers the spike 2 (S2) head finds a receptor known as ACE2, a protein commonly found on the surface of cells. S1 dissociates from the spike and releases the head, which climbs toward the viral capsid and is enveloped by glycans.
Once the probing peptides attach to a cell, the glycans release their hold on S2, which moves like a zipper along a connecting domain to bring the virus into contact with the cell.
Whitford said having better understanding of the mechanism could also help target COVID variants by looking for universal characteristics of the spike. “We’re trying to identify all the relevant moving parts that we might be able to stop,” he said.
That, in turn, should lead to further work by experimental labs and others who wish to build onto the simulation. Whitford noted all the models and data will be freely available to the research community.
“This has been a great experience,” Dodero-Rojas said. “We started this project during my first year of graduate school, and besides how challenging it has been amid the pandemic, the possibility that our work may help fight COVID makes it highly rewarding.”
“I’m optimistic about the next steps,” he said. “Potentially, the mechanism we show could be a target for new therapies or strategies to reduce SARS-CoV-2’s impact on humanity.”
The National Science Foundation (CHE-1614101, PHY-1522550, MCB-1915843, PHY- 2019745), the Welch Foundation (C-1792), the Northeastern University Discovery cluster and research computing staff and the AMD COVID-19 HPC Fund program supported the research.




